CRISPRon
State of the art on-target efficiency predictions for CRISPR-Cas9 based on deep learning utilizing the binding energy model developed for CRISPRoff.

Try the CRISPRon webserver for on-target effiency prediction.
CRISPRoff
Off-target predictions for CRISPR-Cas9 based on an energy model for the RNA-DNA duplex binding. The model out-performs machine learning models on existing off-target data.
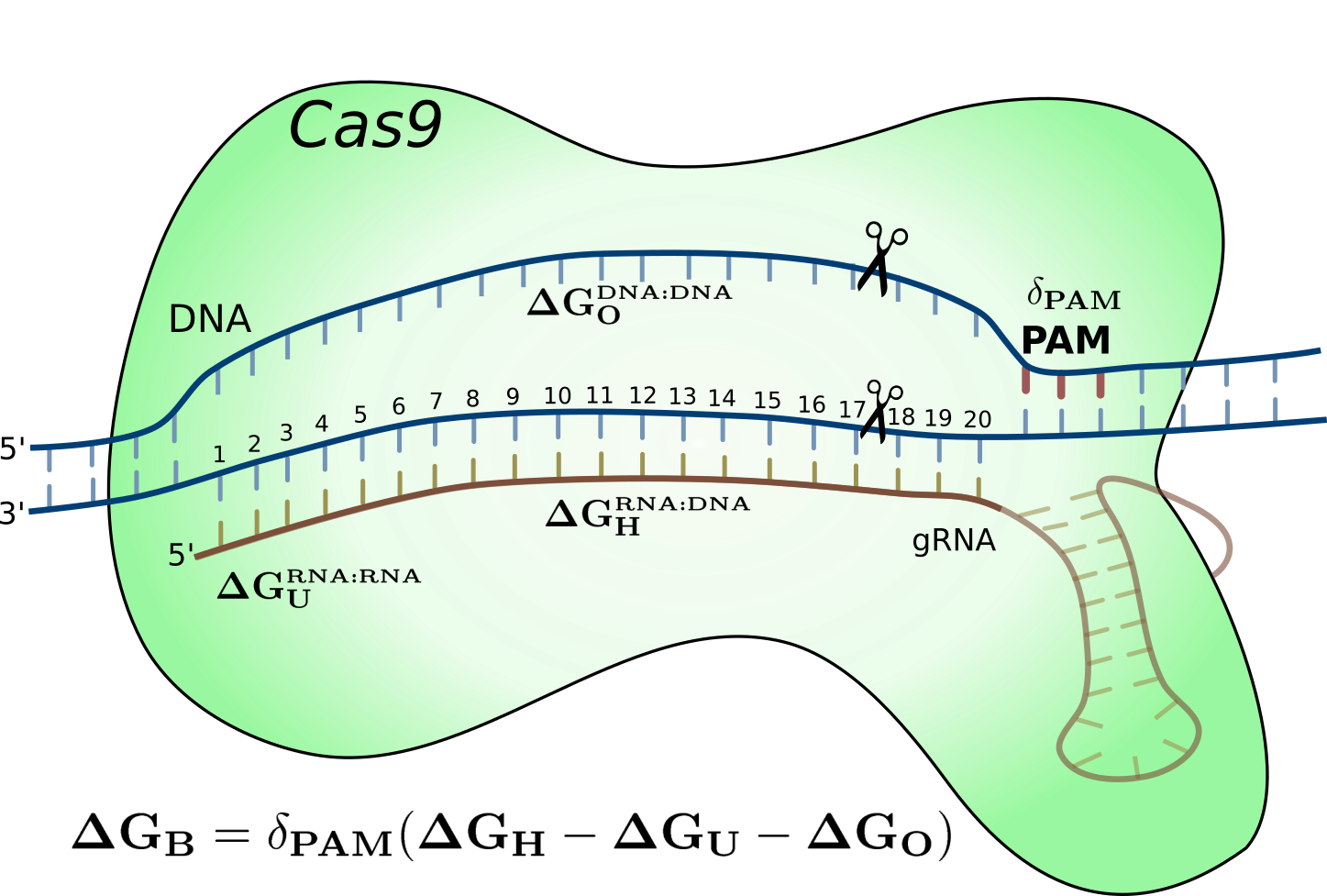
Try the CRISPRoff webserver to predict CRISPR-Cas9 specificity and off-targets.
CRISPRroots
Computational pipeline for the analysis of RNA-seq data from CRISPR/Cas9 edited and control cells. The pipeline offers on-target edit verification and detection of possible off-targets affecting the transcripome.

Download the CRISPRroots pipeline here.
CRISPRon-BE
Design of guide-RNAs for CRISPR/Cas9 basediting for A → G and C → T conversions.

Try the CRISPRon-BE webserver to design guide-RNAs for CRISPR-Cas9 baseditors.
