Webserver for Aligning structural RNAs
Results
| FoldalignM | |
|---|---|
| CPU time: | 217.34 |
| Average sequence ID: | 0.442980500264 |
| Average free energy: | -23.9522580645 |
| Average covariation: | 0.313879456706 |
| Average basepair probability: | 0.698450892452 |
| Canonical basepairs: | NA |
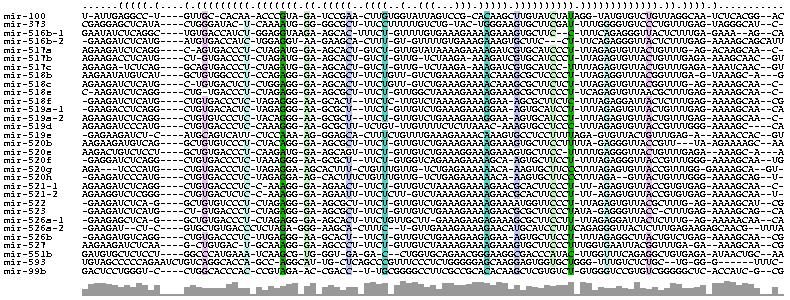
 Sequence and structure consensus for FoldalignM
Sequence and structure consensus for FoldalignM AGAAGAUCYCMKGaucuSYKKKRCCCYYcYHRRRGGvRAcRSCVYUcUUCUbGUUGUYUGAAARAAAAGAAMRBGCYYYYCUyUUWRRRK KKUWMCBKUUKGRGrAAAAGCAAyuCR
......(((((.(....(.(((((((((.(((((((.((.(((((..((((..(..(((...))).))))).)))))))))).))))))) )))))).)).)))).)...........
The alignment colored using colorrna.pl of the Vienna package
Cluster tree of the methods

| Table |
