COAT PhD summer school 2012: RNA-RNA interactions 2012-09-23
made by Stefan E Seemann (seemann@rth.dk)
Lecture slides
Exercise 1:
RNA interactions are fundamentally important to the life cycle of any RNA molecule. Give at least one functional example for each interaction in the following figure! Extend the drawing with more complex interactions!
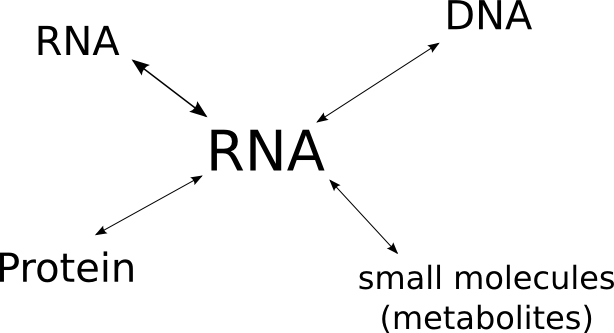
Exercise 2:
Why does so many different predictors exist for RNA-RNA interactions? Find to each of the following figures at least one method that can predict the RNA-RNA interaction!
(a) 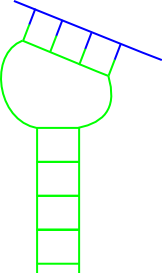 (b)
(b) 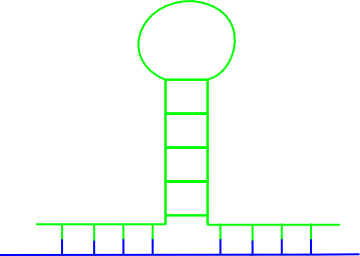 (c)
(c) 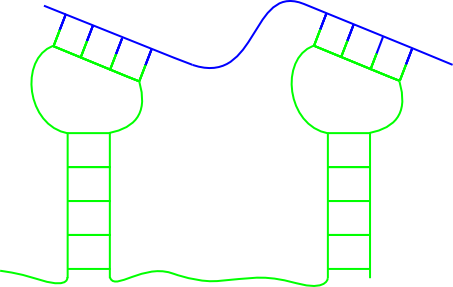
Exercise 3:
(based on Ex.1 of Andreas Richter's EMBO Practical Course on Computational RNA Biology 2010)
In bacteria, small regulatory RNAs (sRNAs) often act as post-transcriptional regulators by base pairing to trans-encoded target mRNAs. The sRNA RyhB interacts, among other targets, with the sodB mRNA.
- Check the sRNA RyhB at the ncRNA database Rfam (http://rfam.sanger.ac.uk/). Where does RyhB bind its target mRNA sodB? Which function does this binding implicate?
- The tool TargetRNA (http://snowwhite.wellesley.edu/targetRNA/) has been designed to search the genome of a species for potential targets of a given sRNA sequence. It evaluates interactions by a Smith-Waterman alignment-like scoring, which is based on base pairing potential instead of homology. Use the TargetRNA web server to identify putative RyhB targets in E. coli.
- The tool IntaRNA (http://rna.informatik.uni-freiburg.de:8080/IntaRNA.jsp) has been designed to predict mRNA target sites for a given non-coding RNA. IntaRNA's scoring is based on hybridization free energy and accessibility of the interaction sites in both molecules. Get the E. coli sequence of RyhB from TargetRNA. Get the E. coli mRNA subsequence of 200 nt length centered around the start codon of sodB (pos. 1733302 to 1733502), f.e. from the UCSC Microbial Genome Browser (http://microbes.ucsc.edu/; select genome 'Escherichia coli K12').
- Compare the predicted RyhB-sodB interactions with the experimental validated interaction from Geissmann and Touati, 2004 (see Figure). How do they differ? In which cases will these methods fail?
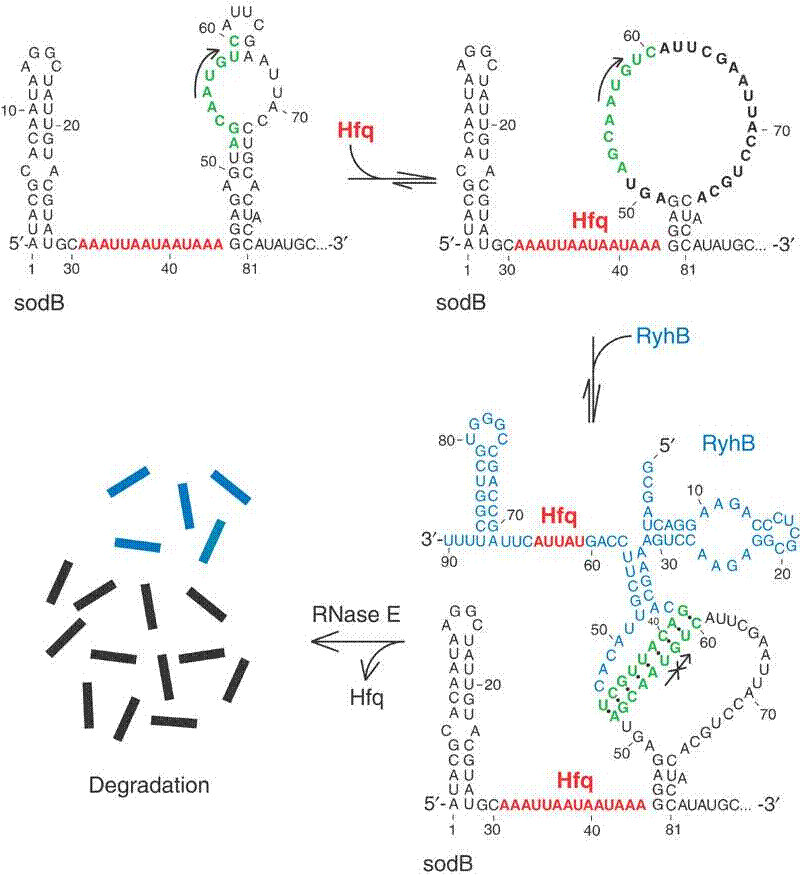
Exercise 4:
Apply DSIR (http://biodev.extra.cea.fr/DSIR/) and RNAxs (http://rna.tbi.univie.ac.at/cgi-bin/RNAxs). Design potent siRNAs to knock down sodB (see Ex.3 if you wonder how to get the sequence)! Compare the predicted siRNAs with the binding site of RyhB! Discuss why nature may have chosen a different post-transcriptional regulator!

 (b)
(b)  (c)
(c) 
