Webserver for Aligning structural RNAs
Results
| MASTR | |
|---|---|
| CPU time: | 111.64 |
| Average sequence ID: | 0.31608005521 |
| Average free energy: | -11.59 |
| Average covariation: | 0.356845238095 |
| Average basepair probability: | 0.49713158679 |
| Canonical basepairs: | NA |
 Sequence and structure consensus for MASTR
Sequence and structure consensus for MASTR umRWDVMMYUSCYUCYSRMASYKRRUWYCMYRDGYUGSYAUMKMWCHGURUWMMWWYUYAYUGKMSWGYMMMAHUYGMWWYCMMMKWSWU RGRSKRMARASHRCS
.........((((((((((((((((((((((((((((.....((...................))...))))))))))))))))).)))) ))))).)).......
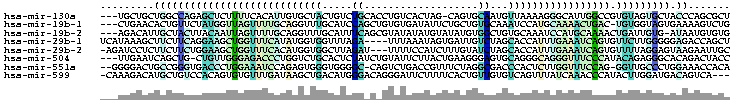
The alignment colored using colorrna.pl of the Vienna package
Cluster tree of the methods

| Table |
