About
RILogo is a software for displaying sequence logos and base pairing between two RNA sequences/alignments. The program is published in: P. Menzel, S.E. Seemann, J. Gorodkin. RILogo: Visualising RNA-RNA interactions, Bioinfomatics, 2012.
Contact
Please contact Peter Menzel (pmenzel@gmail.com) for technical issues, bug reports or general questions about the software.
Download
The latest source code of RILogo can be downloaded from Github. Use git clone git://github.com/pmenzel/RILogo.git to fetch a copy or get the sources as a compressed tar.gz or zip file. RILogo is published under the GNU LGPL 3 licence. Refer to the README file for installation and usage instructions.
Input
The input for the web server is as following:
Structure-annotated Multiple Sequence Alignment
The required input for the web server is a multiple sequence alignment with secondary structure annotation in dot-bracket format. Supported alignment formats are Stockholm, CLUSTAL, or FASTA. The name of the structure annotation line must be structure in Clustal/Fasta format, or #=GC SS_cons in Stockholm format, see examples below.
Example for FASTA format
>gca_bovine GGCCCUGUUUGUGAAUUUACACGUUGAAUUGGGGGCUC&GAAGGCCGGUCAAAUUCAGAUCAAU-CCGGCCA >gca_chicken CACUCUGUAAGUGAAGU-UCAUAAUGAGUUGGGGGUCG&GAUGGCCCACCAAACUCGU-UAUGAGUGGGCCA >gca_mouse AGUCUUAACCGUGAUA-UUCAUGUCGAAUUGGAGAUUU&GGACGUUGGGCAAACUCGAUAAGG-CCCAACGU >gca_rat UUCCUUAAGGGUGAUU-AUCAUGUCGAAUUGAGGGAUA&GGAGGUUGGGCAAACUCGACAAUGUACCAACUA >structure (((.....AAAA......(((......)))....)).)&............((((((..aaaa..)))))).
Example for Stockholm format
# STOCKHOLM 1.0 gca_bovine GGCCCUGUUUGUGAAUUUACACGUUGAAUUGGGGGCUC&GAAGGCCGGUCAAAUUCAGAUCAAU-CCGGCCA gca_chicken CACUCUGUAAGUGAAGU-UCAUAAUGAGUUGGGGGUCG&GAUGGCCCACCAAACUCGU-UAUGAGUGGGCCA gca_mouse AGUCUUAACCGUGAUA-UUCAUGUCGAAUUGGAGAUUU&GGACGUUGGGCAAACUCGAUAAGG-CCCAACGU gca_rat UUCCUUAAGGGUGAUU-AUCAUGUCGAAUUGAGGGAUA&GGAGGUUGGGCAAACUCGACAAUGUACCAACUA #=GC SS_cons (((.....AAAA......(((......)))....)).)&............((((((..aaaa..)))))). //
Example for CLUSTAL format
CLUSTAL W (1.82) multiple sequence alignment gca_bovine GGCCCUGUUUGUGAAUUUACACGUUGAAUUGGGGGCUC&GAAGGCCGGUCAAAUUCAGAUCAAU-CCGGCCA gca_chicken CACUCUGUAAGUGAAGU-UCAUAAUGAGUUGGGGGUCG&GAUGGCCCACCAAACUCGU-UAUGAGUGGGCCA gca_mouse AGUCUUAACCGUGAUA-UUCAUGUCGAAUUGGAGAUUU&GGACGUUGGGCAAACUCGAUAAGG-CCCAACGU gca_rat UUCCUUAAGGGUGAUU-AUCAUGUCGAAUUGAGGGAUA&GGAGGUUGGGCAAACUCGACAAUGUACCAACUA structure (((.....AAAA......(((......)))....)).)&............((((((..aaaa..)))))).
The examples above have both RNAs in one alignment, where the sequences are separated by the '&' character. It is also possible to upload two alignment files separately. In that case, both alignments may have different amount of sequences. Note that the MI is only calculated from the set of sequences which is present in both alignments.
Output
RNA Interaction Logo
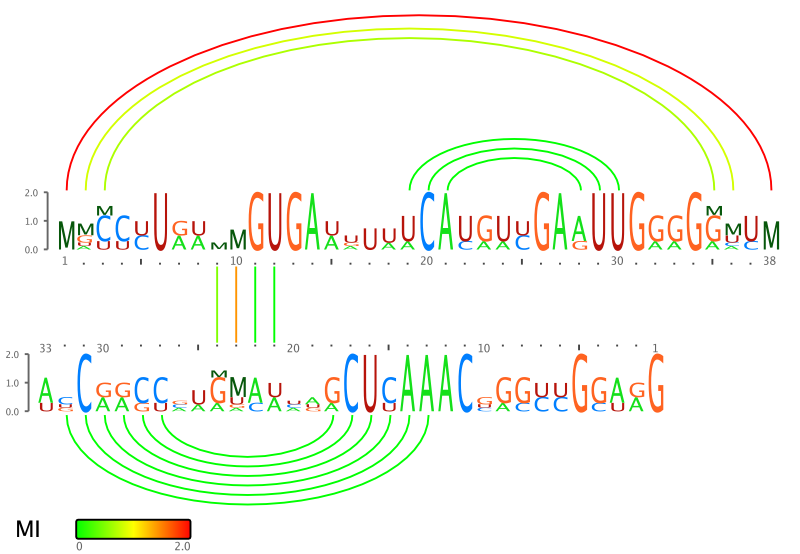
RILogo produces an SVG image with two sequence logos, one for each interaction partner. Intra- and intermolecular base pairs are shown by arcs or straight lines between both logos.
Mutual Information measure
RILogo implements four different measures for the mutual information (MI) of a base pair. The MI is shown by adding the letter M on top of a stack of the letters A,C,G, and U and by coloring the arcs / lines with a color gradient ranging from 0.0 to 2.0 bit. Note, that the letter M is shown on each side of a base pair with half of the MI value in bits, e.g., the M are shown with height 1 bit, when the MI of that base pair is 2 bit. See the supplementary material of the Bioinformatcs paper or this file for the definition of the 4 different MI measures.
Additional options
Upload custom phylogenetic tree
When using treeMI or treeMIWP for calculating the mutual information, RILogo uses the pairwise phylogenetic distances of the sequences in the alignment. RILogo uses fasttree to calculate a phylogenetic tree from the intersection of the sequences in both alignments.
Alternatively, it is possible to upload a custom tree in Newick format, which will be used to extract the pairwise distances between the sequences.
Customize appearence
Example for setting the options no_chars = true and draw_rulers = false.

