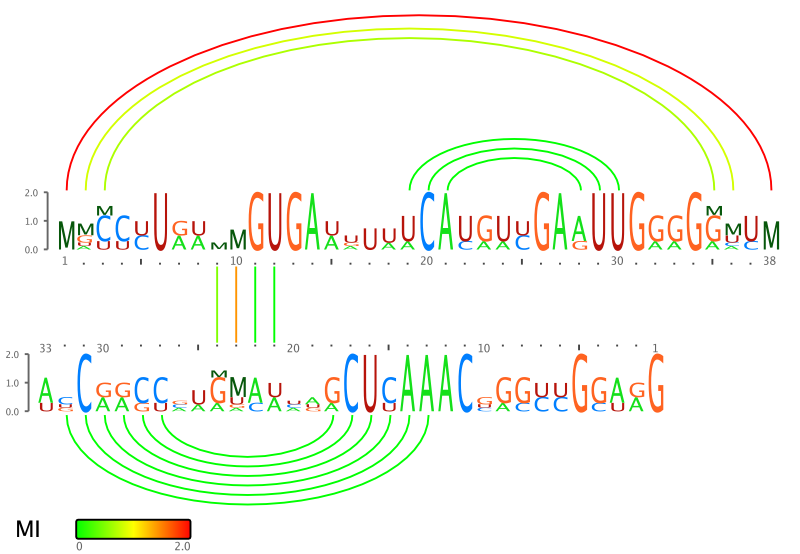
 RILogo creates RNA-RNA interaction logos for two RNAs. The input are
either two single sequences or two multiple sequence alignments with annoation
of intra- or intermolecular base pairing between the two RNAs. RILogo displays
sequence conservation by a sequence logo for each RNA and structure
conservation by the mutual information of the secondary structure base pairings.
RILogo supports four different methods for calculating the mutual information.
RILogo creates RNA-RNA interaction logos for two RNAs. The input are
either two single sequences or two multiple sequence alignments with annoation
of intra- or intermolecular base pairing between the two RNAs. RILogo displays
sequence conservation by a sequence logo for each RNA and structure
conservation by the mutual information of the secondary structure base pairings.
RILogo supports four different methods for calculating the mutual information.
See the Help page for input formats and examples.
RILogo is free of charge and also available for download as an open source standalone program.
Submit
Please fill out the submission form and click the Submit button. Input fields marked with a * are required. (Load Example Data)
