WebCircRNA: Assessing the circular RNA potential of coding and noncoding RNA
Input format
You can either enter BED format input or fasta sequence for multiple candidates or upload a BED format or fasta file, which can be assembled from RNA-seq. The BED input will be predicted using trained model from all 188 extracted features, and the fasta sequence input will be predicted using trained model from only sequence features.
Example-BED
Each candidate must be entered as a single line delimited by tab and must contain 5 columns and the last column must be unique identifier, e.g.:Example-fasta
The sequence must only contain the following four canonical bases 'A', 'C', 'G' , 'T' and 'U'. a FASTA header tag can be specified as the first line that starts with the character '>'. The input sequence must be longer than 100nt, otherwise it will be ignored in our prediction results:Input criteria
(1) The input transcript must be longer than 100 nts.
(2) The input transciprt cannot be longer than 2136788 nts of largetst circRNA in circbase.
(3) The input chromosome must be within 24 chromosomes of human.
(4) For fasta sequence input, We will remove those non-nucleotide sequences, and we will only process the remaining nucleotide sequences (ACGUNT).
(5) For BED format input, the start position must be smaller than end position. The end position cannot be larger than chromosome size .
(6) The names for input transcirpts must be unqiue when you input multiple transcripts.
For those inputs not stastifying the above criteria, they will not be processed by WebCircRNA.
Options
Email options
Optionally, you can specify an email address and a custom job name if you wish to be informed by email when the computation for your job finishes. Otherwise you can use the given job ID to access the results for your job any time later or simply wait until the computation is done. The Results page will be automatically updated every ten seconds until the computation for the job has finished. We recomend this option because it will take some time to finish the computation.
WebCircRNA pipeline
It predict the fractile of being circRNAs for coding and non-coding RNA, which shows the reliability of predicted scores. It integrates 3 models: (i)circRNA potential of PCGs (CP-PCG), it classify circRNA from protein coding genes; (ii) circRNA potential of lncRNAs (CP-lncRNA),the prediction fractiles for classifying circRNAs from lncRNAs in PredcircRNA (Pan et al., 2015); (iii) stem cell potential of circRNAs (SP-circRNA), it predicts stem cell circRNAs from other circRNAs. And provide the link to UCSC circRNA track
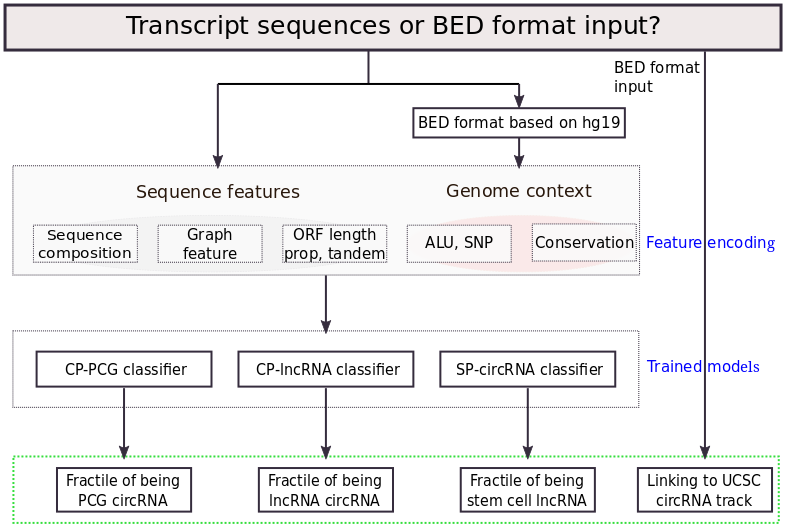
Results
Prediction results
Results are displayed for each entry separately, and The prediction result consists of prediction parts and discovered circRNAs in UCSC browser. For prediction results, one is for predicting circRNA from lncRNAs based on our previous publication, the other two is for predicting circRNA from protein coding genes, and for predicting stem cell circRNA from other circRNAs, respectively. In addition, we also integrate our published circRNA vs lncRNA models for discriminating circRNA from lncRNA
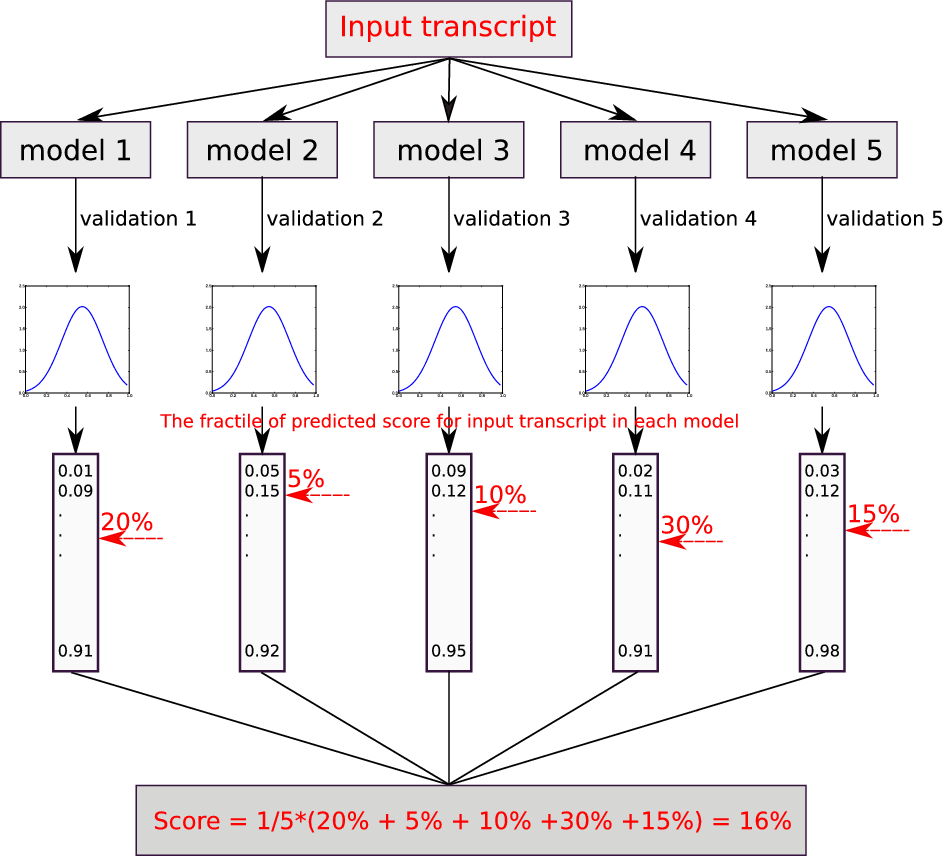
Predicted fractile
We split the training data into 5 subsets, of the 5 subsets, a single subset is kept as the validation data, and the remaining 4 subsets are used as training data.
In our result interface, we calculate the predicted fractiles for input transcripts as follows:
(1) Obtain the predicted probability score distribution each validation dataset, and we got 5 score distributions.
(2) For each score distribution, ranking them in descending.
(3) Predict a score for new input transcript from independent testing dataset, and calculate the fractile of the predicted score in the above 5 score ranking list, respectively, like 10%, 20%.
(4) Average the 5 fractiles for input transcript as final fractile.
Prediction results from circRNA vs lncRNA model CP-lncRNA
It shows the prediction fractile score, from the trained model CP-lncRNA and each line is one gene candidate.
Prediction results from circRNA vs protein coding gene model CP-PCG
It shows the prediction fractile score from the trained model CP-PCG and each line is one gene candidate
Prediction results from stem cell circRNA vs other circRNA model SP-circRNA
It shows the prediction fractile score from the trained model SP-circRNA and each line is one gene candidate.
Discovered circRNAs in UCSC browser
It shows the link to the UCSC browser, which gives experimentially discovered circRNAs in different studies.
Output example
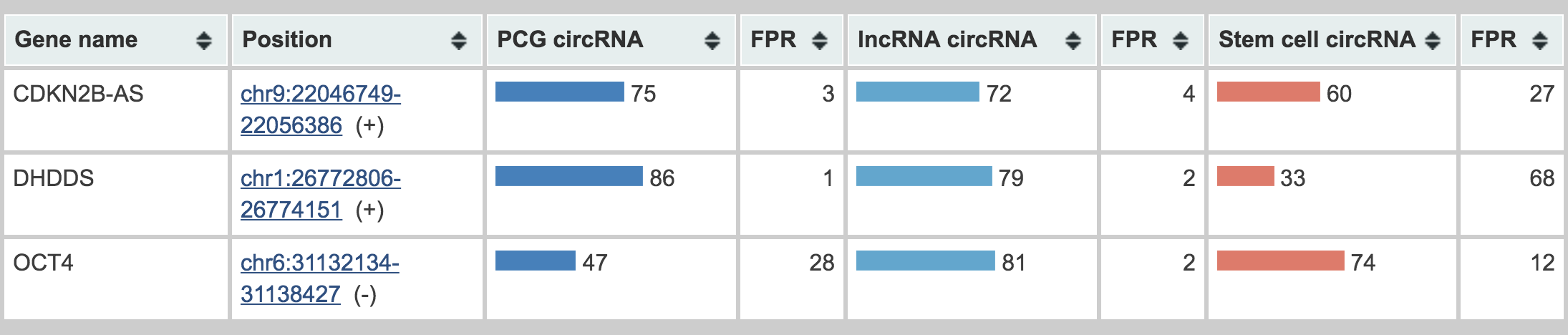
Output from WebCircRNA. Example output for the lncRNA CDKN2B-AS and the two PCGs DHDDS and OCT4 is shown. We thus ignore the “PCG circRNA” score for CDKN2B-AS and the “lncRNAs circRNA” scores for DHDDS and OCT4. Because “stem cell circRNA” scores only apply to circRNAs, this too should be disregarded for OCT4, since it is not predicted to have a circRNA isoform. The respective “FPR” shows the estimated false positive rate of the corresponding methods. When submitting a BED file, the genomic context of any prediction can view in the UCSC browser via the link in the “Position” column.
