|
|
|
|
|
DocumentationContents:
1. IntroductionThis program was written to make editing RNA alignments easy. The package comes with rnadbtool to analyze the alignment and Pfold for predictions. Pcluster is included for clustering sequences into groups with similar secondary structure. FoldalignM is included for aligning RNA structures.
Details can be found in
This program is free software; you can redistribute it and/or modify it under
the terms of the GNU General Public License as published
by the Free Software Foundation; either version 2 of the License, or (at your
option) any later version. This program is distributed in the hope that it will
be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of
MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public
License for more details. You should have received a copy of the GNU General
Public License along with this program; if not, write to the Free Software
Foundation, Inc., 59 Temple Place - Suite 330, Boston, MA 02111-1307, USA.
The application is distributed vith a version for Linux (x86_32) and Mac OS X 10.4 (Intel processor). There is also a source distribution included for other systems.
The requirements of SARSE and the programs toolbox are:
To run the program execute 'SARSE' in the SARSE/bin directory. For easy
access to execution from any directory add a path to your environment
variables:
If you have placed the SARSE directory
on your home directory your path should look like this:
To build the source version do the following: 6. FeaturesSplit viewSplit view splits the screen in two parts (primary and secondary view) that each contains the whole alignment. This easily lets you find pairing or complementary bases by clicking/selecting one/more base(s) in either of the views, e.g. if you click in the primary view, the secondary view will show the pairing/complementary base and vice versa.When working in split mode you can choose between two selection modes - "single select" or "double select". "Single select" only lets you make selections in the primary view, which means that no complementary or pairing bases are shown in secondary view, while "double select" allows selection in both views. SARSE remembers the position from where you run a program from the toolbox. If you're currently showing column 100-150 and you run a program, SARSE will go to that same position when showing the resulting file. This unfortunately does not apply for secondary view in split view. PairingmasksSARSE supports one kind of pairingmasks that consists of any 1-letter code like '1','a' etc. Pairingmasks that contain (, [, {, or > can be used in the double selection mode, but some of the analysis programs does not support this type of pairingmask. The ( and [ can be crossing like in a pseudoknot: ((([[[)))]]].7. Basic Features (howto)Here are presented a small selection of how-to's, describing some common tasks. An overview of all features is presented in the next part Start the programIf you have followed the installation instructions, see doc/install.txt, you can start the editor by simply writing SARSE whereever you are.The program takes 2 commandline parameters (--file and --project). You can start SARSE on the commandline typing: SARSE --file=filenamewhere "filename" is the file you want to use in a new project. Don't use ~ in the path, use absolute paths SARSE --project=projectnamewhere "projectname" is the existing project you want to use SARSE --project=projectname --file=filenameIf you use both parameters together, a new project will be created using the "filename" and the "projectname", and will be located in the current directory. In this way no further clicking is needed to start working. Open file/New ProjectsSARSE is project oriented which means that every time you open a file a project must be created.File -> New This opens a dialog where you are requested to select the file you want to open. When you press 'Open' you are asked for a name of the project and a working directory. Defaults are provided, default projectname is the name of the file without the filetype-postfix(eg. '.col'). Default working directory is the directory from which you started SARSE. You can accept the defaults or give new ones of your liking. If appropiate the file will be copied to the working directory. Example: You start a project with a file named HIV1-leader2.txt then the default project name is HIV1-leader2 Open an existing projectIf you want to open an existing project,File -> Open project Then you get a list of existing projects where you can choose the name of the project you want to open Delete a projectIf you have a project you don't want to work on again delete itFile -> Delete Project Then you get a list of all your projects and you can choose which one to delete. You can only delete 1 project at a time, and it is not allowed to delete an open project. Save file in another location / ExportIf you want to save your alignment outside SARSE e.g. if you wan't to use it in another program or want a fasta fileFile -> Export File Then you select a location, write a filename and select a format. Delete sequencesSelect the names of the sequences you want to delete.Move basesBases can be moved in two ways, mouse or buttons.
Mouse: Moves a selection of bases one position at a time. Buttons: Moves bases within selection from one end to the other. Select area, where you want to move the bases from one side to the other. You can move the bases in the alignment as long as there are gaps to move them into. The whole selection is evaluated together, so if one can't move no one can. Also, it is not allowed to move non-continous blocks of bases. 8. MenusSee also table of menus. File-menu
Edit-menu
View-menu
Tools-menu
Info-menu
9. Alignment editing toolsThis is a series of icons on the left side of the window. Manual editing:
View function:
Pairing function:
Select function:
10. Undoable actionsThe following actions are supported by undo and redo:
The undo/redo history is saved with the project, which means that when reopening a project you can still undo/redo past actions. 11. History windowThe history window consists of both the files created when running Pfold or RNAdbtool and the actions performed on each file. This way you have an overview of the files contained in your project (this does not apply to Pcluster output or files you add to the project directory) and you can always go back to a previous file by clicking on it in the history window. The file currently displayed on your screen is marked with grey and lists all (undoable) operations you have performed on it underneath. If you click an operation it is undone and the font style is changed to italic so you can easily see, which operations you can redo. Basically the history window shows file view combined with a history. When you use keyboard shortcuts to undo/redo these operations are performed on the file currently displayed. Furthermore it is not possible to undo/redo a file that is not displayed. 12. Working with the tableTo mark a single cell, just click on it. To mark an area of cells, hold down the mouse button while you drag it across the table, or mark the first cell and then hold down shift while you expand the area with the arrow buttons on your keyboard. To mark more than one area select one area like above, and then hold down the Ctrl button while you drag the mouse over the next area. Notice that the selected rows always are the same for all selected columns. 13. Programs and pipesIn the Tools menu you can select Programs and a new window opens with a list of programs. You can select any number of programs, but be aware that some programs require execution of other programs. The Programs window provides information about these dependencies, but a rule of thumb is that you can run any RNAdbtool program by itself or combined with other RNAdbtool programs. Pipeline analyses are provided for some program packages instead of including all subprograms in the SARSE toolbox. The directory sarse/programs/pipes/ contains bash scripts that runs a given number of programs as a default analysis. The files produced are saved in a directory and normally a file is returned for SARSE. The available program packages are listed to the left. Click one of the packages. The programs contained in the package are listed on the right with a description. If the program has different options an Option button is displayed next to the program name. If you click the Option button you can adjust the parameters. When done press OK and wait. If any of the programs are sending an error message to the command line a dialog opens and asks if you want to continue. You can choose to see the log message containing the output from the program before continuing. Some programs are sending error messages even though there isn't any fatal error. In any case, view the log message and consider if it looks dangerous. SARSE is currently distrbuted with the following program packages: rnadbtoolsThis is a group of programs used to analyse RNA alignments. The following rnadbtools are available in the SARSE toolbox:
For more information see: http://rnadbtool.kvl.dk. For reference see: Semi-automated update and cleanup of structural RNA databases, J. Gorodkin, C. Zwieb, and B. Knudsen. Bioinformatics, 17:642-645, 2001. PfoldPfold is written by Bjarne Knudsen (bk@daimi.au.dk) and makes secondary structure predictions. For reference see: Knudsen, B. and J. J. Hein (1999) Using stochastic context free grammars and molecular evolution to predict RNA secondary structure. Bioinformatics, 15 (6), 446-454. Pfold runs through a pipe (pfold.pipe) using the following programs:
PclusterThis program clusters sequences into groups with similar secondary structure. For more information see:Semiautomated improvement of RNA alignments E. S. Andersen, A. Lind-Thomsen, B. Knudsen, S. E. Kristensen, J. H. Havgaard, E. Torarinsson, N. Larsen, C. Zwieb, P. Sestoft, J. Kjems and J. Gorodkin, RNA 13:1850-1859, 2007.The Pcluster program is run through the pipe pcluster.pipe and has an option to set the region to be analyzed. Please note that some output from pcluster.pipe does not appear in SARSE, only the resulting col-file is loaded. FoldalignMThis program creates a multiple alignment, and create a common pairingmask for the alignment. The input is unaligned sequences. For further information see the http://foldalign.ku.dk14. Constructing a pipeA special pipes directory is provided: sarse/programs/pipes, that contains default analyses that runs a given number of programs specified in a bash script. The files produced are saved in a directory and normally a file is returned for SARSE. The bash script has a specific header that defines the working directory and the programs directory. If you want to create a pipe of your own for SARSE, then use this header. Below commandline execution of the programs are written and the files are written to the work directory. 15. Adding new programsIn the following we assume you know the basic XML syntax. The only prerequisites that must be fullfilled for a program to be added to SARSE is:
You insert your program immediately after a </program> tag if you are doubt, place it between these 2 tags: </program> </programs> at the end of the file. Program The enclosing tag for each program is a <program> tag it has a few nescessary attributes. Some of them has default values because they are for future extensions. Here is an example from the file: <program name="stem_colors" priority="7" package="coloring tools" selected="false" sequencetype="RNA" type="analyzer" depends=""> </program>The "name" attribute is both the exact name of the command to run the program and the name that is displayed in the menu. "Priority" is for deciding which programs are run first, the lower a number the sooner it is run. The "package" attribute is for grouping the programs in the menu. "Selected" is for a program to be selected by default when you open the program menu. "sequencetype" must be "RNA" and type must be "analyzer", no choice. The "depends" attribute can take the value of the "name" attribute of another program in the xml file. When you select a program in the menu that is dependent on another program that program will also automatically be selected. Description
The program description has a tag of its own and is added like this: <program ...> <programdescription> Colors stems of alignment in different colors. </programdescription> </program> Input-output format
You then need to declare the in- and output formats. At the moment this is limited to col-format so you
have to add the input-formats and output-formats in the following way: <program ....> <programdescription>...</programdescription> <inputformats> <fileextension>col</fileextension> </inputformats> <outputformats> <fileext>col</fileext> </outputformats> </program> Adding commandline optionsIf your program doesn't take commandline options you just insert <parameters/> just before the
</program> and you are done.
<param selected="false" input="false" number="0" spaced=false>Then you add a <name> tag which should be exactly how the option is used on the commandline, including "-" or "--" if used (eg.). <name>-s</name>Then you supply the description <paramdescription> Support information is output as the last entry. </paramdescription>And then end with a closing tag for the option. </param> Another example is an option that takes an input. Then the input attribute of of the <param> must be true. The difference is an additional <input> tag <input number="1" delimiter="" description="limit for support"> 0.75 </input>The "number" attribute tells how many individual values it contains and "delimiter" say what character is used to separate the values. Then there is a "description" attribute and lastly the default-value of the tag. The whole example looks like this: <parameters> <param selected="false" input="true" number="0" spaced="false"> <name>-l</name> <paramdescription> Sets the limit for support. The default is 2/3. </paramdescription> <input number="1" delimiter="" description="limit for support"> 0.75 </input> </param> </parameters> 16. TroubleshootingIf you get: Exception in thread "main" java.util.zip.ZipException: No such file or directory when you start the program, it is because the environment variable SARSE_HOME is not pointing to the SARSE directory. If your jvm runs out of memory e.g. you get a out of memory exception, you can change the maximum memory available in the file bin/SARSE. Look for the line java -Xmx512m -DSARSE_HOME=$SARSE_HOME -jar ../lib/editor.jar. Here you can change the Xmx512m, which means maximum 512 megabytes memory to anything you want. e.g. -Xmx1024m which means maximum 1 gigabytes. 17. Known bugsHere is a list of known bugs sorted according to priority. Please report newly discovered bugs to info@sarse.kvl.dk. Editor bugs:
18. Future wishesPlease send suggestions to info@sarse.kvl.dk. Here is the current list sorted according to priority:
Comments, questions, etc., email
webmaster@sarse.ku.dk. |
| Last updated November 5th, 2007 by E. S. Andersen, A. Lind-Thomsen and J. Gorodkin |
 = Change Symbol. Change the marked symbols to
any symbol in the alphabet. This function is also used for editing the
pairingmask.
= Change Symbol. Change the marked symbols to
any symbol in the alphabet. This function is also used for editing the
pairingmask. = Split. Splits the current view in two.
The behaviour of the two windows are such that the upper/left window is
the
= Split. Splits the current view in two.
The behaviour of the two windows are such that the upper/left window is
the = Unsplit. Unsplits the current
view.
= Unsplit. Unsplits the current
view. = Unpair Bases: The selected bases,
and the bases they pair with, are all shown in lower case.
= Unpair Bases: The selected bases,
and the bases they pair with, are all shown in lower case. = Pair Bases: The selected base names
are changed to upper case if they are within the pairingmask, and a gc,
au or gu base pair
= Pair Bases: The selected base names
are changed to upper case if they are within the pairingmask, and a gc,
au or gu base pair = Pair Columns: Expands the
pairingmask. Asks for a letter/number/parenthesis to use in the
pairingmask, and then pairs all cg,au and gu basepairs in the two
columns. Works on any equal number of columns. Pairing functions: For
all three pairing functions it is the case that when the letters are
changed to upper case, the pairings for each sequence is resolved,
which means that you have no guaranties that the bases will actually
pair when the operation is done.
= Pair Columns: Expands the
pairingmask. Asks for a letter/number/parenthesis to use in the
pairingmask, and then pairs all cg,au and gu basepairs in the two
columns. Works on any equal number of columns. Pairing functions: For
all three pairing functions it is the case that when the letters are
changed to upper case, the pairings for each sequence is resolved,
which means that you have no guaranties that the bases will actually
pair when the operation is done.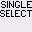 = Single Select. Only the selected bases are selected, not the counterparts.
= Single Select. Only the selected bases are selected, not the counterparts. = Double Select. Selects not only the selected bases, but also the bases the pair with.
= Double Select. Selects not only the selected bases, but also the bases the pair with.