RAIN: RNA–protein Association and Interaction Networks (v1.0)
RAIN integrates non-coding RNA (ncRNA) and protein interaction networks in an easily accessible web interface.It contains three types of ncRNA associations: microRNA-target, ncRNA-protein and ncRNA-ncRNA interactions and combines them with protein-protein interaction available in the STRING database. ncRNA associations cover four model organisms and are extracted from experimental data, automatic literature mining and curated examples. For miRNAs, we further include precomputed target predictions.
RAIN developers
RAIN is being developed at CBS, CPR, RTH, SIB, DTU, KU and UZH.
RAIN reference
RAIN: RNA-protein Association and Interaction Networks
Junge A, Refsgaard JC, Garde C, Pan X, Santos A, Alkan F, Anthon C, von Mering C, Workman CT, Jensen LJ*, Gorodkin J*
Database (Oxford). 2017 Jan 10;2017:baw167. Print 2017
[ PubMed | Paper | Webserver ]
Contact
Alexander Junge ajunge@rth.dkXiaoyong Pan panxy@rth.dk
Searching in RAIN
ncRNA identifiers
For miRNA we expect miRBase identifiers or accession numbers; for other ncRNAs we accept Refseq, Ensembl as well as identifiers from the main databases of the respective organism as follows:human: HGNC,
mouse:MGI,
rat:RGD,
yeast: SGD.
The ncRNA alias mapping file used in RAIN is available from our Download page.
Protein/mRNA identifiers
For all organisms covered, RAIN accepts the same identifiers and accession for proteins as STRING. Furthermore, RAIN collapses proteins and mRNAs coding for the respective protein into a single node to integrate pre- and post-translational interactions. This means that, e.g., miRNA-mRNA and protein-ncRNA interactions involving the corresponding mRNA and protein will result in two edges adjacent to a shared protein/mRNA node.
Step 1. Input query:
You can input a single ncRNA or protein identifier and choose an organism of interest to search against RAIN. You can input a single ncRNA or protein identifier and choose an organism of interest to search against RAIN.
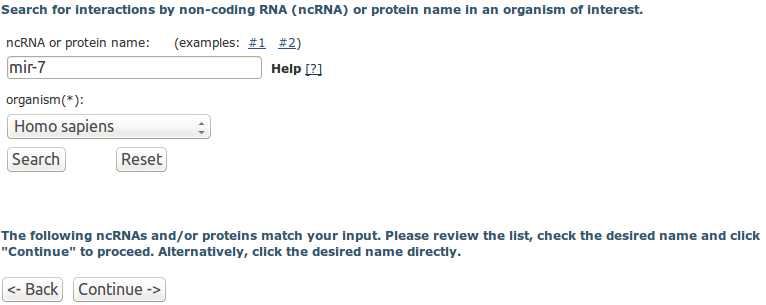
Step 2. Search result:
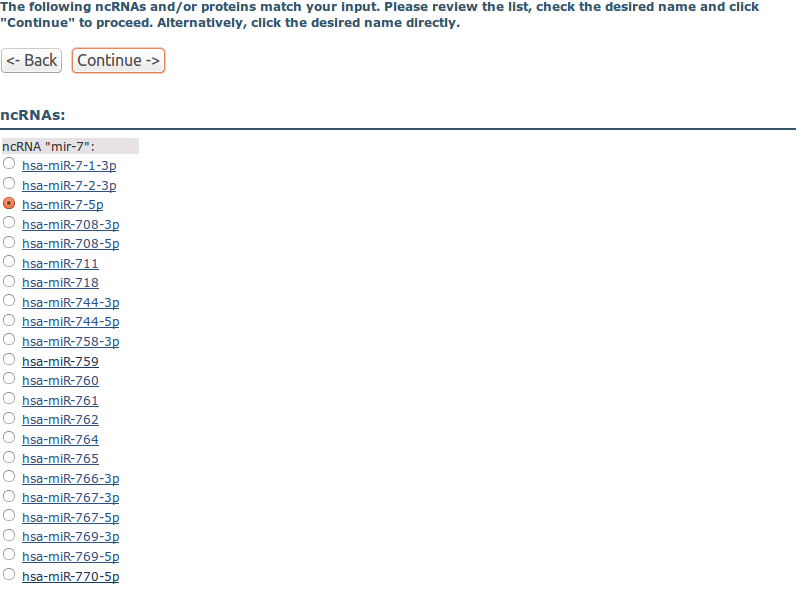
In the next step, the desired ncRNA or protein identifier is chosen:
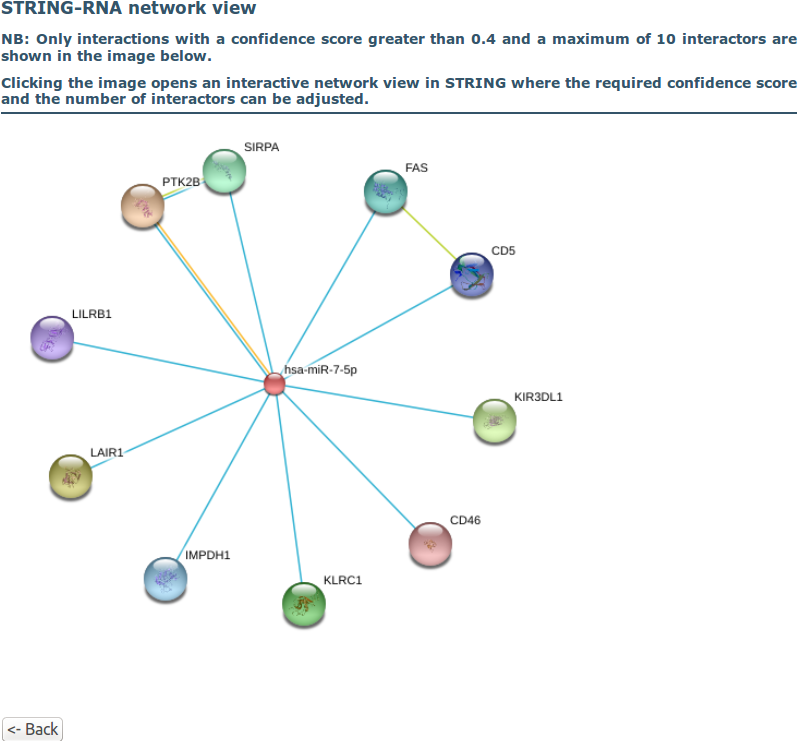
Step 3. Interaction network:
Finally, the interaction network for the input identifier is displayed. Clicking the network links to an interactive version of the network.
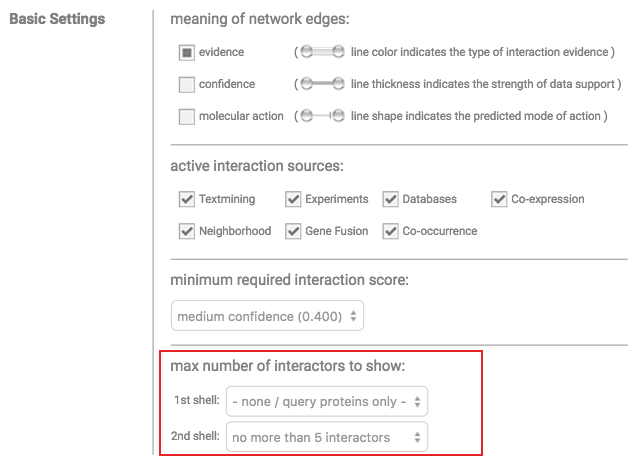
The interactive network view, provided by STRING, allows to select specific evidence channels to be displayed. This can be achieved by clicking 'Data Settings' below the network visualization and then checking/unchecking the tick boxes for each evidence channel under 'active interaction sources:' (see image below).
Note that selected evidence channels are stored between session.
This may make edges in RAIN networks appear to be missing.
In this case, please adjust the active evidence channels.

Viewing interaction information and text mining evidence:
Information about each interaction can be accessed by clicking the edge in the interactive network view. This opens a popup window displaying confidence scores, split by evidence channel. Detailed information are available by clicking the 'Show' button in the popup window.
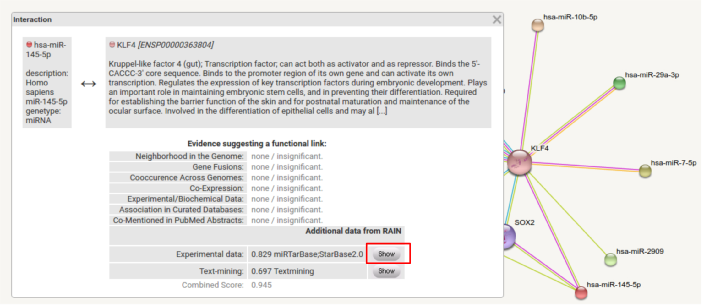
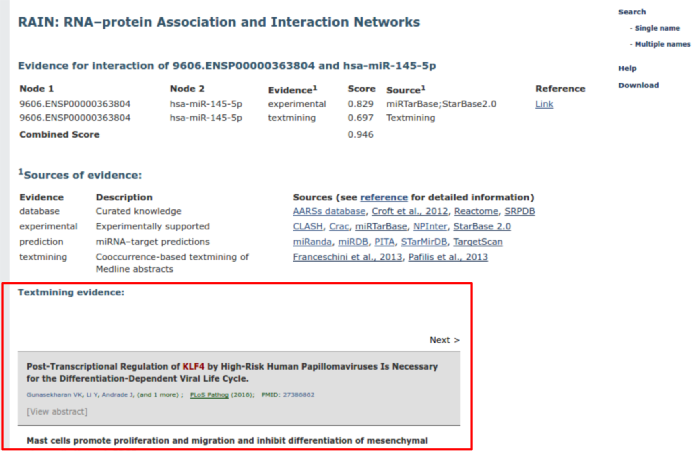
Despite listing confidence scores and sources for each interaction, detailed text mining evidence is displayed if the interaction is supported by automated minining of the scientific literature.
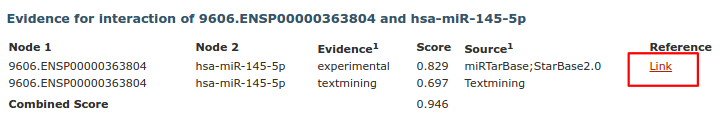
If an interaction is supported by experimental evidence, a link to a PubMed search for the articles describing the experiments is displayed (if avaiable).
Extended multiple names search:
RAIN's multiple names search only returns interactions between the queried entities by default.
Interactions adjacent to the queried entities can be found by adding a second shell of
interactions in the Settings tab of the interactive network view: