CRISPRoff (v1.2beta): off-targeting assessment of Cas9 gRNAs
About
Background information for the CRISPRoff tool
CRISPRoff and CRISPRspec scores
CRISPRoff scores are computed for individual off-target predictions of a gRNA. It corresponds to the approximate free energy gain of the gRNA:off-target binding. Higher free energy gain hints to more efficient cleavage of the off-target, therefore, higher CRISPRoff scores correspond to higher confidence for individual off-targets, predicted for a specific gRNA. Our extensive benchmarks show that CRISPRoff score is more accurate than other off-target scoring methods when predicting cleaved off-target regions.
CRISPRspec score represents the specificity of the selected gRNA/on-target sequence, based on the predefined off-target landscape (e.g. whole genome). It can be used to compare the off-targeting potential of different gRNA/on-target selections. Higher specificity corresponds to lower off-targeting potential, which could also hint to efficient cleavage of the intended on-target.
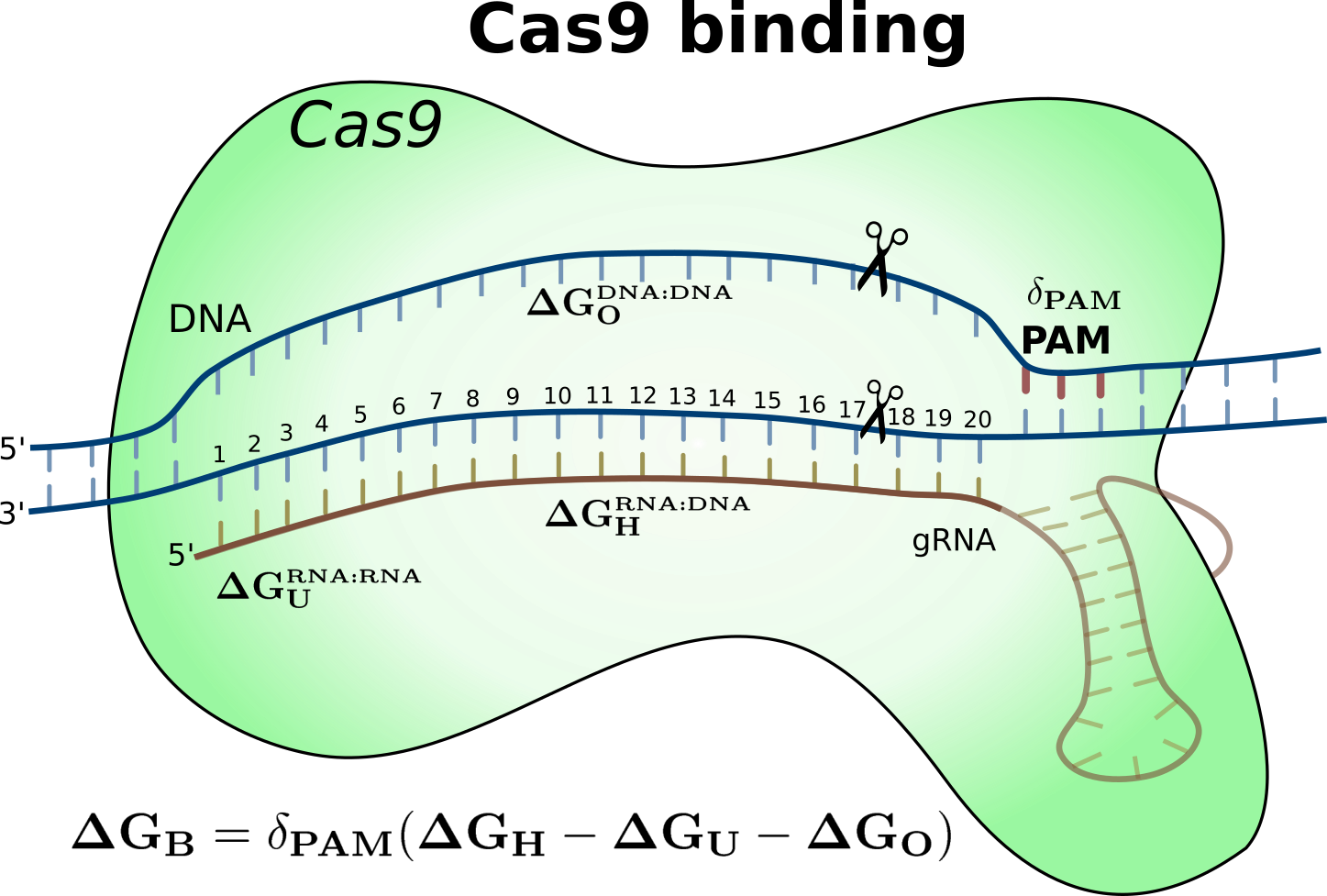
Both of these scores are dependent on our approximate free energy model for
Cas9:gRNA:DNA binding. The full details of the energy model (shown below) and
how CRISPRoff and CRISPRspec scores are computed, can be found in the
reference publication that is given at the bottom of this page (Alkan et
al., 2018).
How to interpret the result page of the webserver
When your job is finished, we present the off-targeting assessment results in a
table format summarizing all given or generated gRNAs that could target given
sequence(s). gRNAs with multiple on-targets appear multiple times in the table
together with the unique genomic coordinates of every on-target.
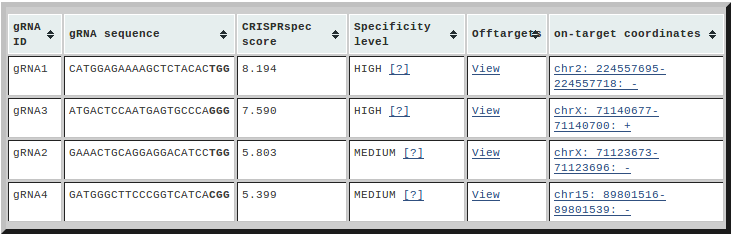
In the table, first column contains the IDs given to the gRNAs under
consideration; second column contains the on-target (gRNA+PAM) sequence where
the PAM is highlighted; third column shows the computed CRISPRspec scores of
the gRNAs; fourth column corresponds to the specificity level of the gRNAs
based on their CRISPRspec scores and the fifth column redirects you to
CRISPRoff result files (individual off-target predictions of each gRNA). Note
that the CRISPRspec thresholds for different specificity levels of gRNAs are
organism-specific and the values for human are taken from our publication
(Alkan et al., 2018) that assigns 89234 unique gRNAs into 3
similar-sized specificity groups; "LOW", "MEDIUM" and "HIGH". In addition,
gRNAs with no off-target predictions are tagged as "MAX" specificity. If the
off-target predictions were performed by the webserver, we also present the
genomic coordinates of all gRNA on-targets (chromosome:start-end:strand) in the
sixth column. By clicking the on-target coordinates, you can open the UCSC
browser in new window where all on-targets and off-targets are uploaded as
custom track and the focus is on the genomic coordinates given in this column.
Downloadable CRISPRoff result files are specific to every gRNA under
consideration, and they contain the target predictions of that unique gRNA. The
webpage displays the 50 highest scoring offtargets but you can also access all
off-targets by clicking the link on the bottom of the top results page.
Additionally, you can download all off-targets as a tsv (tab separated values,
can be imported in excel) file.
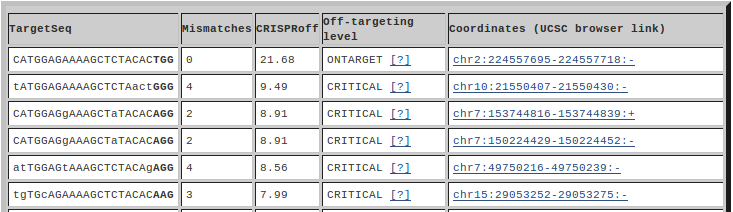
In the html tables shown here, the first column is the target sequence where lower case letters indicate mismatches and PAM sequences are always given in bold. The second column is the number of mismatches between the target and the gRNA. Note that mismatches in the PAM sequence are not counted. The third and fourth columns are the off-targeting score and level, see below. When off-target predictions are performed by the webserver, the final column shows the genomic coordinates of the target. By clicking the genomic coordinates, you can open the UCSC browser in new window where all on-targets and off-targets are uploaded as custom track and the focus is on the genomic coordinates given in this column.
The thresholds for the off-targeting level are based on the ROC analysis in (Alkan et al., 2018) focusing on the Hauessler benchmark dataset (Haeussler et al., 2016). All perfect matches are labeled as ON-TARGET whereas off-targets are binned into 4 groups; CRITICAL, MAJOR, MODERATE and MINOR; where binning thresholds are selected as CRISPRoff scores that corresponds to 0.01, 0.05, and 0.1 FPR within the performed ROC analysis. A CRITICAL off-target has a CRISPRoff score higher than ~6.92, the minimum value that would result in 0.01 FPR when predicting the off-targets of the Hauessler benchmark dataset. If an off-target has a CRISPRoff score lower than this value but higher than ~2.26, the value for 0.05 FPR, it is tagged as MAJOR off-target. MODERATE off-targets have CRISPRoff scores lower than ~2.6 and higher than ~-0.22, the 0.1 FPR. All other off-targets are tagged as MINOR off-targets and these thresholds are the same for all organisms.
Running on organisms besides Human
The CRISPRoff method has only been benchmarked against off-target data from CRISPR-Cas9 experiments performed in human cell lines. Extending the method to other organisms is straight forward, but since off-target data used in the benchmarking was from human only at the time of publication, the application of CRISPRoff to non-human experiments should be treated with a bit of extra caution.
References
CRISPRon/off: CRISPR/Cas9 on- and off-target gRNA design
Anthon C, Corsi GI, Gorodkin J*
Bioinformatics. 2022 Dec 13;38(24):5437-5439
[ PubMed | Paper | Webserver ]
CRISPR-Cas9 off-targeting assessment with nucleic acid duplex energy parameters
Alkan F, Wenzel A, Anthon C, Havgaard JH, Gorodkin J*
Genome Biol. 2018 Oct 26;19(1):177
[ PubMed | Paper | Webserver | Software ]
RIsearch2: suffix array-based large-scale prediction of RNA-RNA interactions and siRNA off-targets
Alkan F*, Wenzel A*, Palasca O, Kerpedjiev P*, Rudebeck AF, Stadler PF, Hofacker IL, Gorodkin J*
Nucleic Acids Res. 2017 May 5;45(8):e60
[ PubMed | Paper | Software ]
Feedback
We greatly appreciate your comments. Open Feedback form in a new tab. Alternatively you can E-mail us with your questions and comments.
