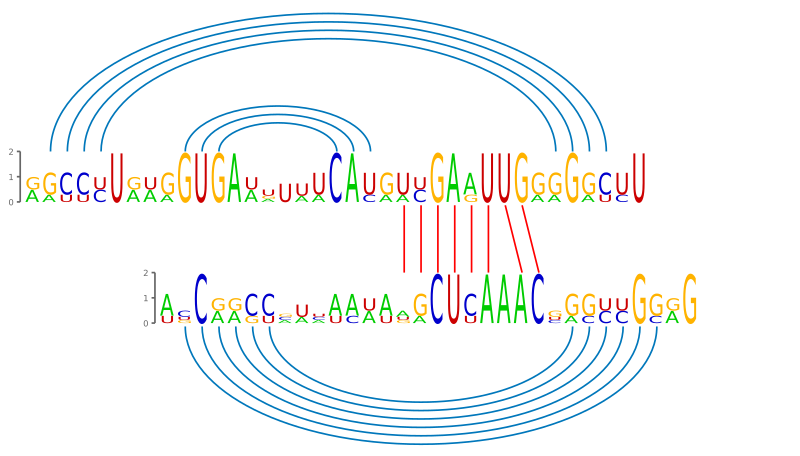
This toy example illustrates the input and output of the PETcofold web server.
Input
RNA alignment
The two alignments in FASTA format look like this, download the alignment
files:
example1.fasta, example2.fasta.
>gca_bovine AGCCCUGUGGUGAAUUUACACGUUGAAUUGGGGGCUU >gca_chicken GACUCUGUAGUGAAGU-UCAUAAUGAGUUGGGGGUCU >gca_mouse GGUCUUAAGGUGAUA-UUCAUGUCGAAUUGGAGACUU >gca_rat AGCCUUAAGGUGAUU-AUCAUGUCGAAUUGAGGGCUU |
>gca_bovine GAGGCCGGUCAAAUUCAGAUCAAU-CCGGCCA >gca_chicken GAGGCCCACCAAACUCGUUUAA-AGUGGGCCA >gca_mouse GGCGUUGGGCAAACUCGAAAAAU-CCCAACGU >gca_rat GGGGUUGGGCAAACUCGAAAAUCUACCAACUA |
Note that the identifiers in both alignments must be the same.
Phylogenetic tree (optional)
The phylogenetic tree in Newick format looks like this:
((gca_bovine:0.078102,gca_chicken:0.41034):0.345797,gca_rat:0.024156,gca_mouse:0.165124):0.001;
Note that the names in the tree must match the names in the alignment file.
RNA secondary structure of duplex (optional)
The RNA secondary structure of duplex in dot-bracket format looks like this:
.((((.................[[[[[[[[.))))..&..((((((.]].]]]]]].......)))))).
Note that the length of the structure has to be the same as for the concatenated alignments.
Output
The output shows the input, the phylogenetic tree, the PETcofold plain text output including the predicted joint RNA secondary structure and score, an image of the covariance information in the joint RNA multiple alignment, and a dotplot of base pair and single stranded reliabilities calculated by the PETcofold scoring scheme:Joint phylogenetic tree
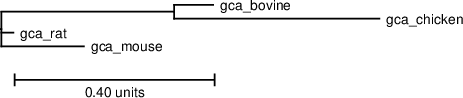
Download PS | PDF | Newick format
PETcofold output
1st step - intra-molecular folding using PETfold
2nd step - inter-molecular cofolding
Download full output (verbose mode)Predicted consensus joint RNA secondary structure
Dotplot of PETcofold reliabilities of base pairs and single stranded positions
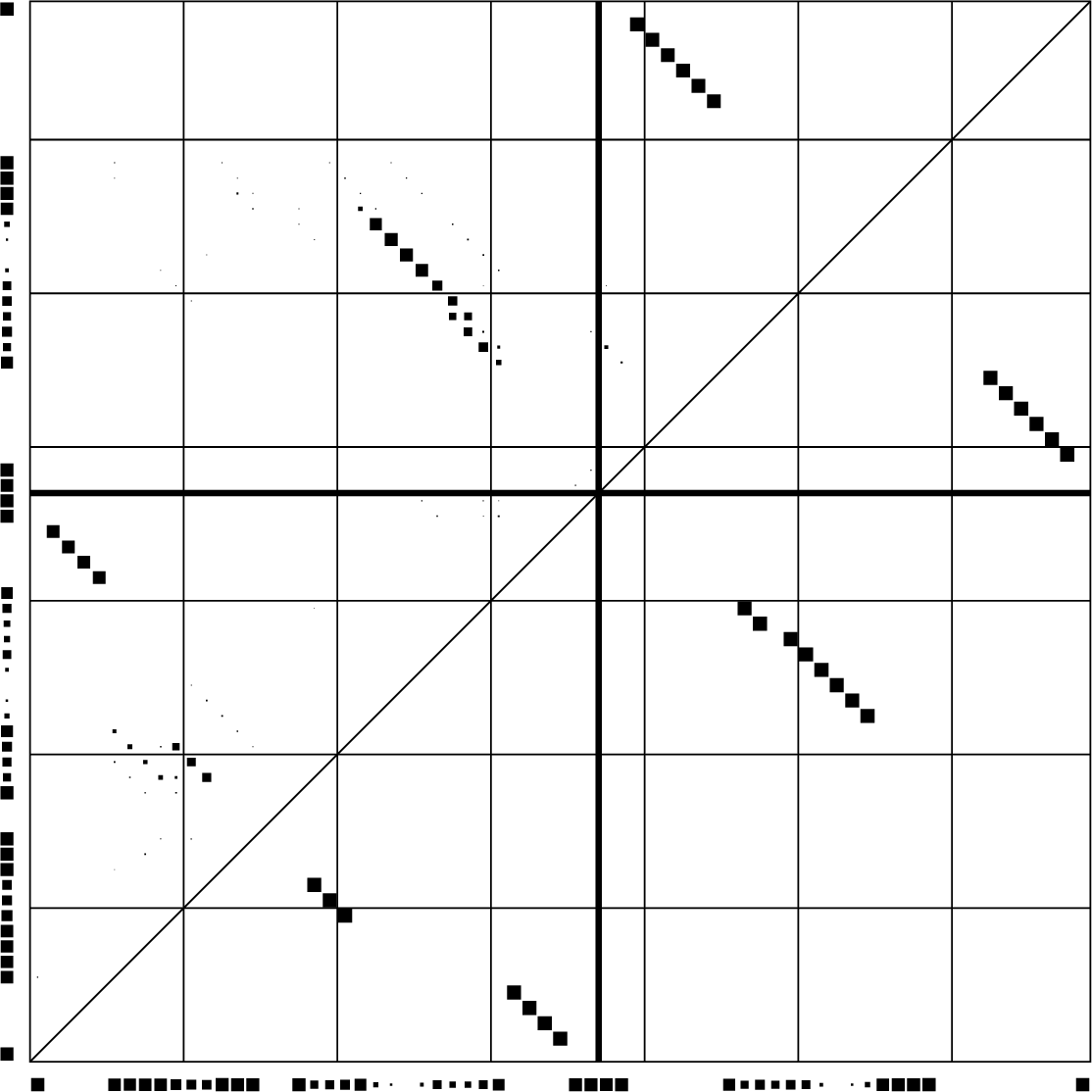
Download PS | PDF | dotplot file