This example illustrates the usage of an Rfam seed alignment for
predicting the RNA-RNA interaction in vertebrates between snoRNA
mgU6-77 and U6 snRNA presented in Seemann SE et.al.,
Bioinformatics, 2010.
An example of two customized alignments, you will find here.
Input
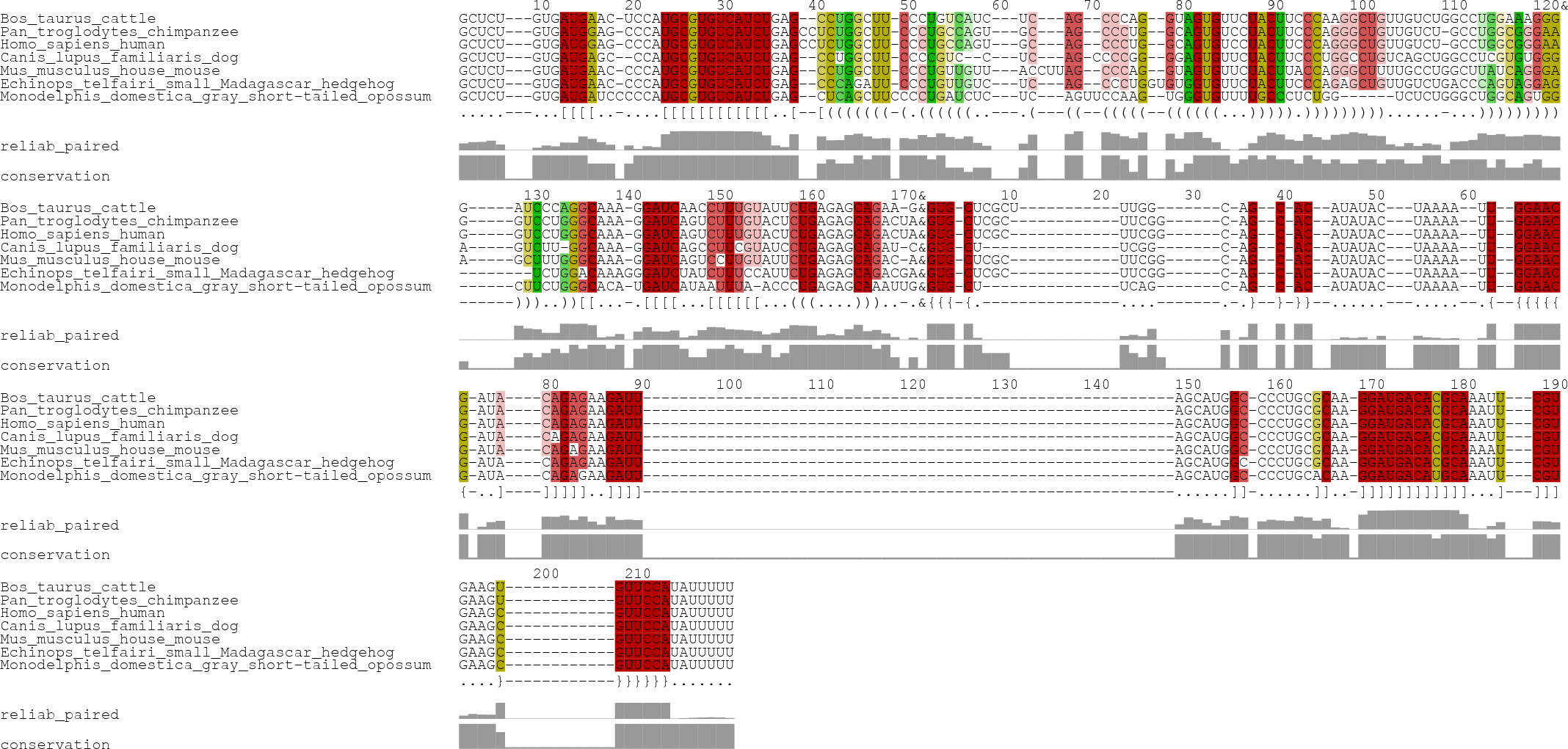
RNA alignment
The Rfam seed alignment of the snRNA U6 can be chosen from the drop down list in the input form. The alignment of the snoRNA mgU6-77 was fetched from snoRNABase and its species names (identifier) were adapted using the drop down list of U6 identifiers: mgU6-77.fasta.
>Homo_sapiens_human GCUCU---GUGAUGGAG-CCCAUGCGUGUCAUCUGAGCCUCUGGCUU-CCCUGCCAGU---GC---AG--CCCUG--GCAGUGUCCUACUUCCCAGGGCUGUUGUCU-GCCUGGCGGGAAG-----GUCCUGGGCAAA-GGAUCAGUCUUUGUACUCUGAGAGCAGACUA >Pan_troglodytes_chimpanzee GCUCU---GUGAUGGAG-CCCAUGCGUGUCAUCUGAGCCUCUGGCUU-CCCUGCCAGU---GC---AG--CCCUG--GCAGUGUCCUACUUCCCAGGGCUGUUGUCU-GCCUGGCGGGAAG-----GUCCUGGGCAAA-GGAUCAGUCUUUGUACUCUGAGAGCAGACUA >Mus_musculus_house_mouse GCUCU---GUGAUGAAC-CCCAUGCGUGUCAUCUGAG--CCUGGCUU-CCCUGUUGUU---ACCUUAG--CCCAG--GUAGUGUUCUACUUACCAGGGCUUUUGCCUGGCUUAUCAGGGAA-----GCUUUGGGCAAA-GGAUCAGUCCUUGUAUUCUGAGAGCAGAC-A >Bos_taurus_cattle GCUCU---GUGAUGAAC-UCCAUGCGUGUCAUCUGAG--CCUGGCUU-CCCUGUCAUC---UC---AG--CCCAG--GUAGUGUUCUACUUCCCAAGGCUGUUGUCUGGCCUGGAAAGGGG-----AUCCCAGGCAAA-GGAUCAACCUUUGUAUUCUGAGAGCAGAA-G >Echinops_telfairi_small_Madagascar_hedgehog GCUCU---GUGAUGAAC-CCCAUGCGUGUCAUCUGAG--CCCAGAUU-CCCUGUUGUC---UC---AG--CCCUGGUGUGGUGUUCUACUUCCCAGAGCUGUUGUCUGACCCAGUAGGAG--------UCUGGACAAAGGGAUCUAUCUUUCCAUUCUGAGAGCAGACGA >Canis_lupus_familiaris_dog GCUCU---GUGAUGAGC--CCAUGCGUGUCAUCUGAG--CCUGGCUU-CCCCGUC--C---UC---AG-CCCCGG--GGAGUGUUCUACUUCCCUGGCCUGUCAGCUGGCCUCGUGUGGGA-----GUCUU-GGCAAA-GGAUCAGCCUUCGUAUCCUGAGAGCAGAU-C >Monodelphis_domestica_gray_short-tailed_opossum GCUCU---GUGAUGAUCCCCCAUGCGUGUCAUCUGAG--CUCAGCUUCCCCUGAUCUC---UC---AGUUCCAAG--UGGGUGUUUUGCCCUCUGG------UCUCUGGGCUGGCAGUGG------CUUCUGGGCACA-UGAUCAUAAUUUA-ACCCUGAGAGCAAAUUG |
Note that the identifiers in both alignments must be the same.
Options
The option Constraint stems get extended by inner and outer base pairs was chosen.
Output
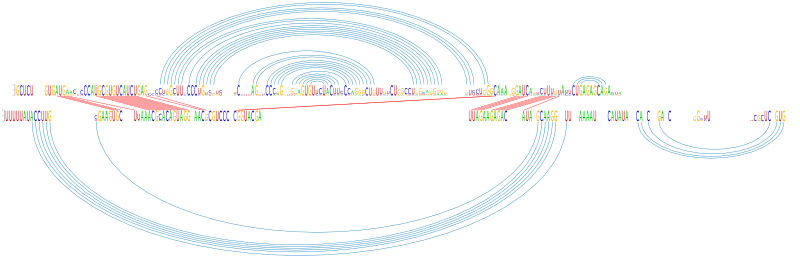
The output shows the calculated phylogenetic tree, the PETcofold plain text output including the predicted joint RNA secondary structure, score and relative score, an image of the sequence conservation in the joint RNA multiple alignment, and a dotplot of base pair and single stranded reliabilities calculated by the PETcofold scoring scheme:Joint phylogenetic tree
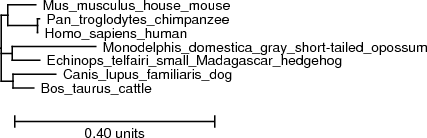
Download PS | PDF | Newick format
PETcofold output
1st step - intra-molecular folding using PETfold
2nd step - inter-molecular cofolding
Download this filePredicted consensus joint RNA secondary structure
Dotplot of PETcofold reliabilities of base pairs and single stranded positions

Download PS | PDF | dotplot file