News
Current news
CRISPR Cas9 base editing efficiencies

Excited that the collaboration with Yonglun Luo lab on base editing gRNA design has been published in Nature Communications. We generated efficiency and outcome data for ~20000 gRNAs and employed deep learning learning models trained on ours and publicly available data by labeling the individual data sets resulting in a cutting-edge base editor design tool. See the paper here.
Previous news
CRISPR/Cas9 efficiency explored: binding energies and PAM context

30 May 2022. Researchers from RTH and Aarhus University describe the energy-based binding mechanisms of CRISPR/Cas9-gRNAs and use an energy model to assess the cleavage efficiency of Cas9-gRNAs. The model is further applied to describe the DNA "sliding" of Cas9 competing for overlapping PAMs. Read the article here or see the news flash on CRISPR medicine news.
CRISPRroots
2021 December 15. RTH researchers construct new pipeline for post CRISPR-Cas9 assessment of on- and off-targets by combining CRISPR off-target analysis with RNA-seq data and variance calling. The software and the paper are available. The paper was highlighted on CRISPR Medicine News.
CRISPRon

2021 May 28: In joint collaboration with researchers from Aarhus University, Lars Bolund Institute of Regenerative Medicine and Harvard Medical School, we present CRISPRon for enhanced gRNA efficiency prediction by deep learning based on generated and publicly available data. Read the full article in Nature Communications and visit the webserver. Also featured on CRISPR Medicine News.
CRISPR-Cas9 energy model
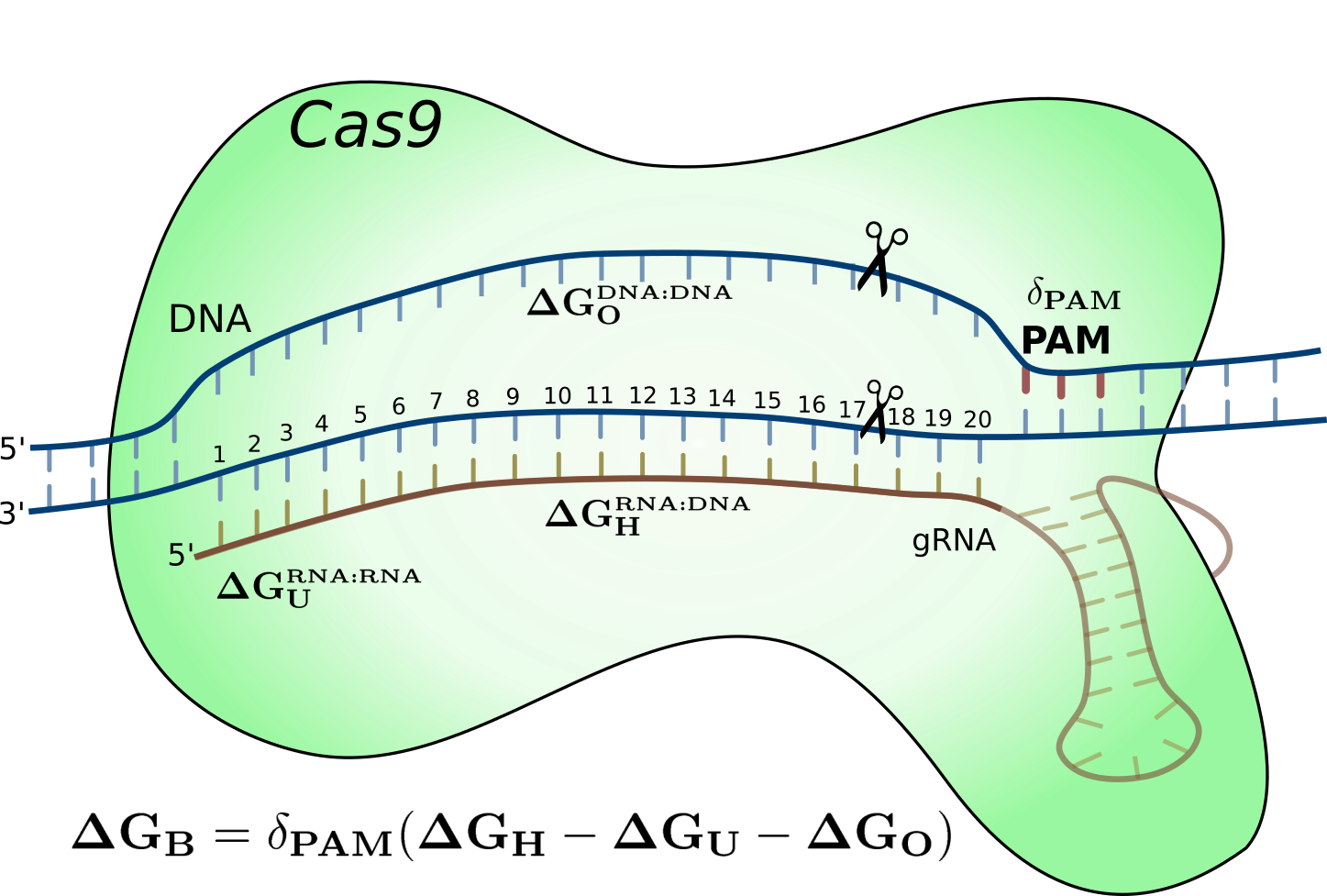
2018 Oct 26: We present a novel energy model for the CRISPR-Cas9 system by modeling the nucleotide duplex binding energy. This model account for a portion of the off-target data from NGS applications as well as perform better than machine learning models. We also provide a specificity measure for how well a gRNA is on-target versus off-target. Read the full article or visit the webserver .
Conserved RNA structure in the dark matter

2017 May 14: RTH researchers discover ~500,000 conserved RNA structures (CRSs) in the dark matter of the genome by using an RNA structural alignment strategy of the vertebrate genomes. The discovered CRSs locate near known genes, regulatory regions and protein binding sites. Expression studies, especially a CRS targeting CaptureSeq experiment in human fetal brain, were employed to characterize the CRSs further. Read the CRS paper here .
RISearch2
2017 Jan 20: RTH publishes a new RNA-RNA interaction prediction method, RIsearch2, that can perform large-scale screens with an exceptional runtime. RIsearch2 together with its post-processing pipeline is a state-of-the-art method for transcriptome-wide prediction of siRNA off-targets. Read the RISearch2 paper here .
Transcriptomic landscape of lncRNAs in inflammatory bowel disease
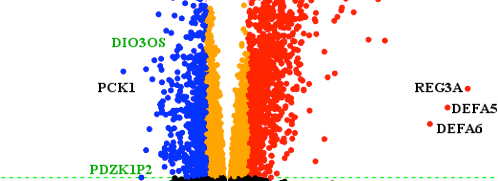
2015 May 13: Researchers lead by Prof Flemming Pociot from RTH publish the first paper on genome-wide transcriptome profiling of human long non-coding RNAs(lncRNAs) and protein-coding genes in Inflammatory bowel disease (IBD). The identified lncRNA transcriptional signature and clinically parameters suggest a biomarker potential in IBD. Read the paper titled " Transcriptomic landscape of lncRNAs in inflammatory bowel disease" .
Profiling Ribose Methylations in RNA

2014-11-26: Researchers lead by Prof. Henrik Nielsen from RTH publish the first paper on profiling Ribose Methylations in RNA based on high-throughput sequencing. Whereas high-throughput methods has now been established for DNA, searches for RNA by high-throughout technologies is just emerging. Here the authors describe their method, RiboMeth-Seq, for discovery of one of the most abundant types of modifications in RNA. Read the paper titled "
Profiling of Ribose Methylations in RNA by High‐Throughput Sequencing
"" .
RNA bioinfomatics book
2014 Mar 25: New book on RNA computational and bioinformatic methods
List of contents and link to the publisher .
cphRNA 2014: Summer School in RNA Bioinformatics
RTH and COAT are co-organizing a Summer School in RNA Bioinformatics. The event will take place on June 23rd to June 28th in the Copenhagen area. Details to follow. Program and application procedure .
Serarching for miRNAs by their transcriptomic profiles
2013 Jul 29: RTH publish a new type of search for microRNA genes. In constrast to more traditional search strategies using the composition of the genomic DNA sequence, the strategy here directly aiming at exploiting the characteristic "finger print" microRNAs leave transcriptomic (RNA-seq) data. The search tool is available via the mirdba webserver . The work is described in our recent paper: " MicroRNA discovery by similarity search to a database of RNA-seq profiles ".
RTH contributes to the RNA annotation of the porcine genome
2012 Nov 15: Through joint national and international collaborations RTH contributes to the annotation of the porcine genome, constituting one of the largest efforts in annotation of noncoding RNAs (ncRNAs) in a single go in a released mammalian genome sequence.
The annotation was conducted by searching for similarity to RNAs from other organisms taking the structural aspect of RNA into account followed by careful semi-automatic curation on selected subsets of the data. In the porcine genome the curation procedure reduced the number of candidate RNA loci from 3,601 loci to 1,976. A particularly interesting example is the class of yRNAs, part of a molecular complex involved in quality control in the cell. It is found in four closely situated copies in mammalian genomes, but in the porcine genome we found that the sequence of one of the four copies were altered to a degree were it is unlikely to be active.
Official press release for the pig genome
Press release from University of Copenhagen
Download the porcine genome paper from Nature
Fellowship to Aashiq Hussain Mirza
2012-05-03: RTH PhD student Aashiq Hussain Mirza has been awarded the Poul and Erna Sehested Hansens Foundation Fellowship of 100,000 DKK.
PhD Summer Course - apply now
2012 Mar 02: RTH is co-organizing a PhD course on RNA sequencing and computational analysis together with Centre for Computational and Applied Transcriptomics (COAT). The course is held at Dept. of Biology, University of Copenhagen on August 20-24, 2012. Topics to be covered are parallel sequencing technologies, microRNA biology and -targeting, protein-RNA interactions, RNA structure an antisense oligonucleotide drugs. The course is a mixture of lectures introducing the topics and hands on computational excercises. No prior programming experience is required, the course aims at PhD students and postdocs within the field of Molecular Biology with an interest to learn how to use bioinformatic tools. After participating in the course, you will be able to do fairly advanced analysis yourself. The course will finish by a symposium with presentations from distinguished international lectures. Updates regarding the course will be posted on rna.dk . Fill out this Registration Registration form and send it to Katrine Mohr . For more information summer school program here .
SME grant from the Strategic Research Council
2011 Jan 21: The Center has received a supplementary grant of DKK 600 000 from the Strategic Research Council. The aim of this funding is to invite small and medium-sized enterprises to collaborate on a project entitled "Discovery of regulatory non-protein-coding RNA genes".
The overall goal of this initiative from the Strategic Research Council is to facilitate short-term involvement of new business partners in research projects. This means that small or medium-sized enterprises, who may not have the resources to engage in long-term projects, are still offered the opportunity to participate in business and society related research.
For the business enterprises the outcome may be tangible and immediately usable results, but an incentive for participating may also be the opportunity to build up new channels of knowledge - to the benefit of the growth and development of the enterprises in the long run.
For the research institutions the collaborative projects offer an opportunity to bring new knowledge and experience into the projects, ie. to have theories or methods tried out in concrete scenarios, to get new inspiration from practice, and/or include special competencies from the business world.
From a societal point of view it is important to strengthen ties between research and business. It adds more relevance to research, creates a basis for innovation, and strengthens the development of a knowledge-based business world. This is the driving force behind the Strategic Research Council's initiative.
Companies interested in further information may contact Center Director Jan Gorodkin at phone +45-3533 3578 or at gorodkin@rth.dk latest by 6 May 2011.
Center for non-coding RNA in Technology and Health (RTH) opens.
2010-05-01: The Center for non-coding RNA in Technology and Health (RTH) opens.
For current news, see here



