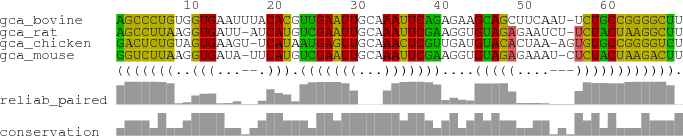
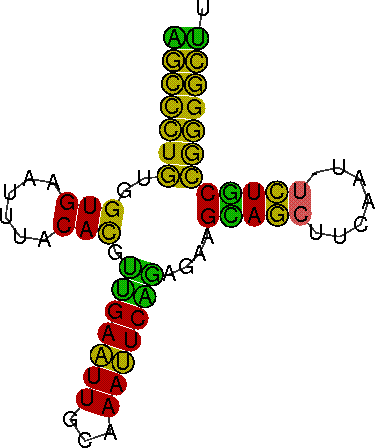
This example illustrates the input and output of the PETfold web server.
Input
RNA alignment
The alignment in FASTA format looks like this, download the alignment file: example.fasta.
>gca_bovine AGCCCUGUGGUGAAUUUACACGUUGAAUUGCAAAUUCAGAGAAGCAGCUUCAAU-UCUGCCGGGGCUU >gca_chicken GACUCUGUAGUGAAGU-UCAUAAUGAGUUGCAAACUCGUUGAUGUACACUAA-AGUGUGCCGGGGUCU >gca_mouse GGUCUUAAGGUGAUA-UUCAUGUCGAAUUGCAAAUUCGAAGGUGUAGAGAAAU-CUCUACUAAGACUU >gca_rat AGCCUUAAGGUGAUU-AUCAUGUCGAAUUGCAAAUUCGAAGGUGUAGAGAAUCU-UCUACUAAGGCUU
Phylogenetic tree (optional)
The phylogenetic tree in Newick format looks like this:
(gca_chicken:0.37262,(gca_rat:0.017848,gca_mouse:0.070927):0.290067,gca_bovine:0.15964):0.001;
Note that the names in the tree must match the names in the alignment file.
RNA secondary structure (optional)
The RNA secondary structure in dot-bracket format looks like this:
(((((((..(((......))).(((((((...)))))))....(((((.......)))))))))))).
Output
The output shows the input, phylogenetic tree, the PETfold plain text output including the predicted RNA secondary structure and score, images of the predicted consensus RNA secondary structure, and a dotplot of base pair and single stranded reliabilities calculated by the PETfold model:Phylogenetic tree
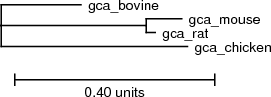
Download PS | PDF | Newick format
PETfold output
Download this filePredicted consensus RNA secondary structure
Dotplot of PETfold reliabilities of base pairs and single stranded positions
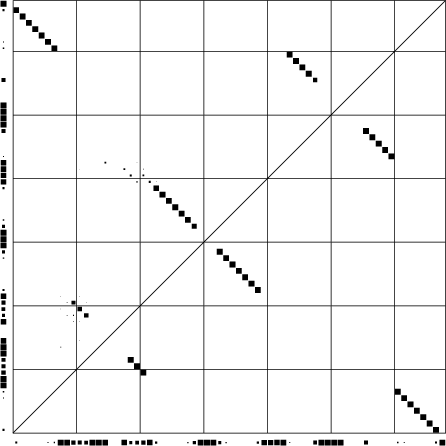
Download PS | PDF | dotplot file