BSGatlas download options:
1. Excel workseet
The summary BSGatlas annotations can be downloaded in a single excel file. This file contains genome coordinates for all genes (both coding and non-coding), UTRs, transcripts, TSSs, and terminator structures. This file also includes of the list of operons.
2. GFF3
The Generic Feature Format Version 3 (GFF3) is a standardized file format for genome annotations. This file format can be read by the IGV and IGB browser applications. The standard should allow straightforward use of our annotation in most -omics analysis workflows. We provide our annotation relative to the Bacillus subtilis 168 annotation as decribed by the The annotation coordinates are relative the NCBI assembly reference ASM904v1.
Simplified. We also provide a simplified version of the GFF files, that does not contain TSS, Terminator, and Operons entries. In particular the latter are implied by transcript informaiton that is still contained.
Download:
- BSGatlas GFF (version 1.1)
- BSGatlas GFF, simplified (version 1.1)
- BSGatlas GFF (version 1.0)
- Genome sequence
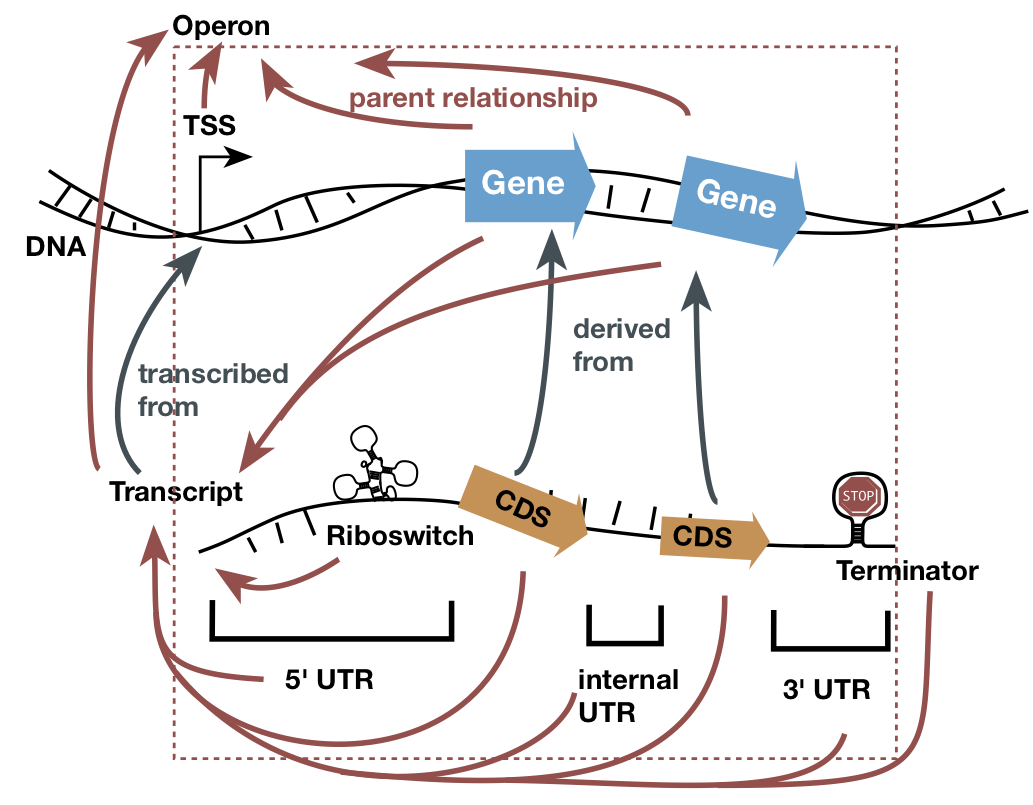
Graphical presentation of the official GFF3 representation of a bacterial
operon. Each operon/UTR/gene/structure is a row in the file, although each gene
also has an extra entry to represent the transcribed region. The relationships
between the entries are notes as indicated by the arrows:
The GFF file itself a tab separated table file. Lines starting with '#' are comments. The columns in the file are:
- Chromosome: Here `ncbi168` for coherence with the browser hub
- Source: Is BSGatlas for this annotation.
- Type: The type for an annotation feature
- Start: Start and ...
- End: ... ending position of a feature.
- Score: Here not used, constant dot as value
- Strand: Strand of a feature. Possible values are {"+", "-", "."}
- Phase: Translation offset phase for coding sequences. Is either "0" or ".".
- List of attributes
Attibutes are a semicolon separated list of
Example: Name=GeneA;Synonyms=GeneX,GeneY
The following keys are used:
| ID: | Unique identifier for each row |
| Parent: | List of hierarchical parent relationships |
| Derives_from: | Link to gene from which a transcript feature originates |
| Name: | The name of a feature |
| locus_tag: | A locus identifier for genes; this often is the used identifier in external databases. |
| Description: | Free descriptive text of a feature |
| comment: | more ellaborate descriptive text of a feature |
| synonyms: | List of alternative names |
| go: | List of gene function terms accodting to the Gene Ontology |
| ec: | List of Enzyme Classifications |
| subtiwiki_category: | A gene classification system from SubtiWiki |
